Research
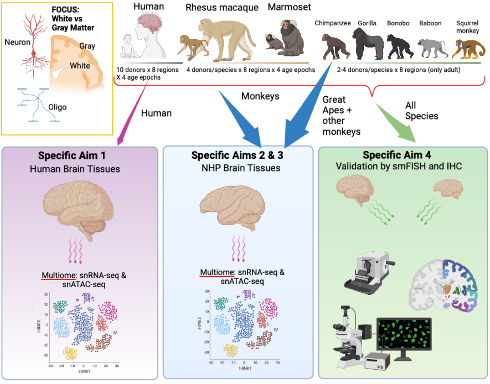
Neurogenomics of the Human Brain Evolution
Using post-mortem human and non-human primates brains, we will assess the developmental trajectories of the gray/white matter in multiple region of interests. Our group will take advantage of single cell multiomics. In specific, our lab will analyze single cell expression and chromatin profiles from corpus callosum, optic chiasm, pyramidal tract, and cerebellar white matter in human and non-human primates across several developmental epochs that match currently funded BICAN grants. We are interested on the oligodendrocyte evolution and these comparisons are key because oligodendrocyte maturation varies dramatically between gray and white matter with white matter oligodendrocytes maturing much earlier in development. Computational methods will be used to define developmental trajectories and to integrate the multi-omics data processed with the aim to refine and prioritize coding and non-coding regions important for human brain development and evolution. By integrating GWAS risk-loci and common variants associated with disorders and complex traits, we will provide the association of neurogenomics of the human specific signatures with neuropsychiatric disorders at coding and non-coding level.
Investigating the role of the autism risk gene CHAMP1 in human iPSC-derived neurons
Patients with CHAMP1-associated NDD are characterized by intellectual disability, autism spectrum disorder (ASD)-like behaviors, microcephaly, hypotonia, and dysmorphic features. In our lab we are leveraging human in vitro models, immunohistochemistry, electrophysiology, and single cell multiomics to determine how CHAMP1 mutations modulates neuronal maturation and proliferation programs relevant to NDD and ASD. The combination of these approaches can generate the data required to understand the function of CHAMP1 in neurons and NDD with the translational potential to inform next-generation therapeutics.


Evolutionary Genomics
Our group will take advantage of cells and tissues from human and non-human primates to characterize the molecular mechanisms under evolution on the human lineage. Our group will take advantage of induced pluripotent stem cells (iPSC) from multiple great apes. Combining RNA-seq and ATAC-seq, we will uncover the human-specific evolutionary trajectories relevant for the maturation and proliferation of neuronal and glia derived cells. We will further take advantage of this in-vitro system as a platform for gene modeling. Integration with GWAS-risk loci will be performed to assess whether human-specific trajectories are affected by loci that put at risk for specific neuropsychiatric disorders.
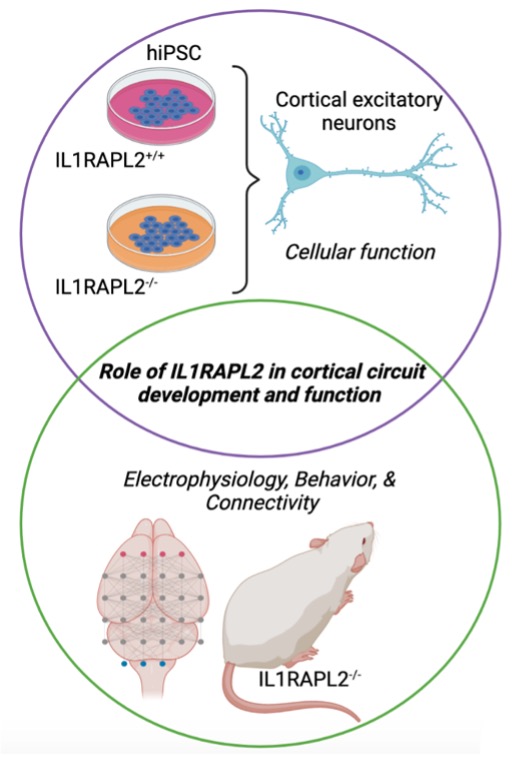
The role of NDD risk gene, IL1RAPL2, in cortical neuron development and function
Mutations in IL1RAPL2 are linked to increased risk for Autism Spectrum Disorder (ASD) and intellectual disability (ID). In our recently published human subjects’ imaging genomic study, we found that IL1RAPL2 expression was highly correlated with memory oscillatory patterns measured by intracranial electroencephalography (iEEG), and it showed high connectivity (e.g., Hub) in a co-expression module specifically associated with this firing frequency. Importantly, genes co-expressed with IL1RAPL2 mRNA are linked to synaptic transmission and are enriched for ASD risk loci and genes whose expression is altered in ASD. We will use a multidisciplinary, cross-species approach to explore IL1RAPL2 functions in cortical development.
